Recent strides in multimodal large language models (MLLMs) have opened up new avenues in medical decision-making. However, many of these models suffer from practical limitations, such as excessive computational costs and task-specific rigidity.
To overcome these challenges, researchers have introduced Med-MoE (Mixture of Domain-Specific Experts) a lightweight, versatile model designed to tackle both generative and discriminative medical tasks like visual question answering (VQA) and image classification.
Med-MoE achieves high performance with only a fraction of the parameters required by other models, offering a significant reduction in computing costs while maintaining accuracy and interpretability.
Challenges in Medical Multimodal Models
Traditional MLLMs such as LLaVA, MiniGPT-4-V2, and CogVLM excel in general multimodal tasks but falter when applied to the medical domain. These models are typically trained on web data, which doesn't align well with the complexity and specificity of medical information.
Furthermore, the high number of parameters, running into billions required by models like LLaVA-Med makes them impractical for many clinical settings, especially those with limited computational resources. While domain-specific models like Med-Flamingo and Med-PaLM have shown promise, they remain focused on a narrow set of tasks and are often too resource-intensive for real-world applications.
Med-MoE addresses these gaps by combining efficiency with versatility, making it a powerful tool for various medical tasks in resource-constrained environments. This article will break down how Med-MoE works, its unique architecture, and its applications in the medical field.
Architecture of Med-MoE
Med-MoE's architecture is built around three key phases: multimodal medical alignment, instruction tuning and routing, and domain-specific MoE tuning. This modular approach allows the model to effectively process different types of medical data, such as images and text, while minimizing computational load.
GitHub repository for Med-MoE.
Phase 1: Multimodal Medical Alignment
The first phase involves aligning medical images—such as CT scans, MRIs, and X-rays—with corresponding language model tokens. A vision encoder extracts tokens from the medical images, which are then paired with text descriptions to ensure that the model can correctly interpret and describe medical images.
This alignment process is essential for multimodal models that must understand both the visual and textual aspects of medical information. By doing so, Med-MoE gains the ability to handle diverse input types and produce accurate, clinically useful insights.
Phase 2: Instruction Tuning and Routing
In the second phase, Med-MoE enhances its ability to follow medical instructions through instruction tuning. The model is trained using a dataset of medical queries and responses, allowing it to excel at multimodal tasks such as answering medical questions based on image data.
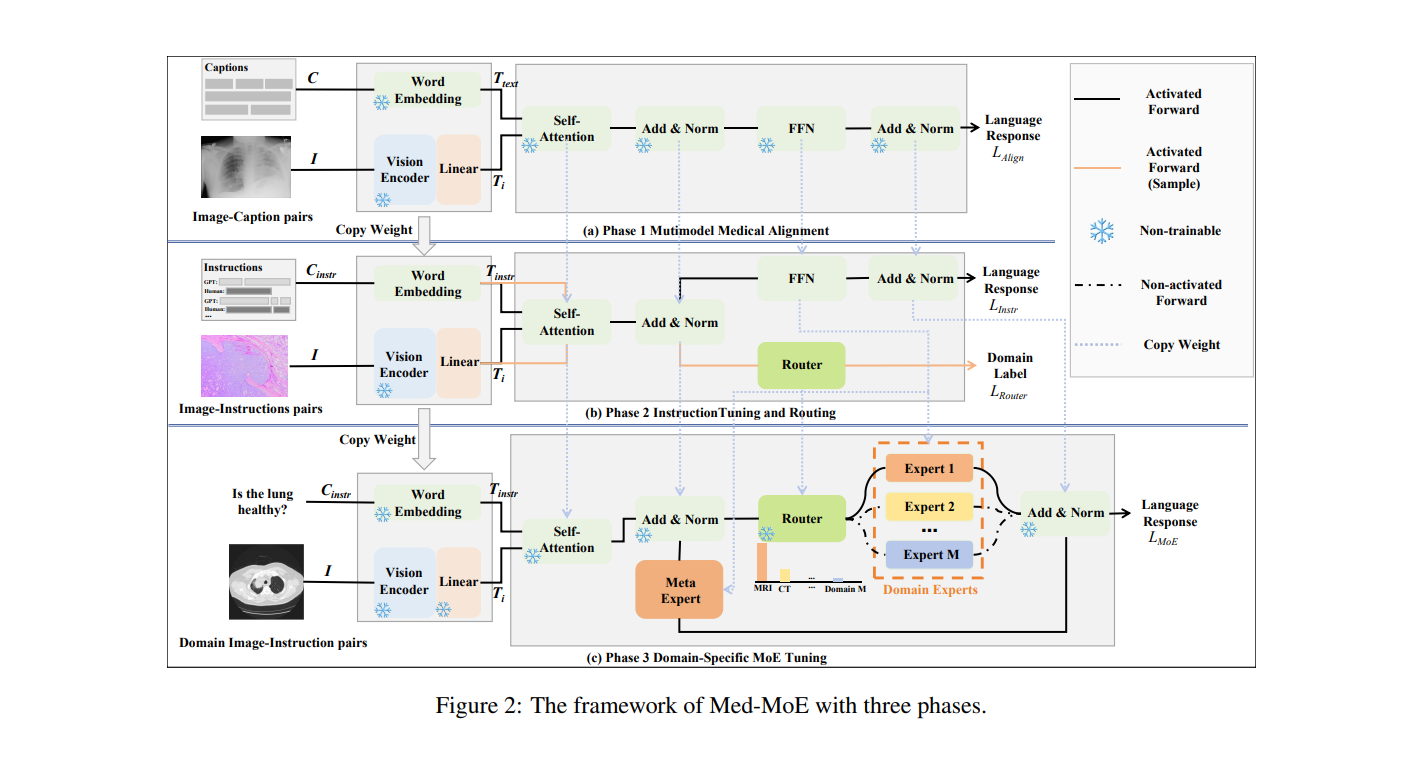
A key innovation here is the routing mechanism, which uses a specialized router to assign tasks to different domain-specific experts based on the input data's modality (e.g., MRI, CT scan). This approach mirrors the collaborative nature of medical diagnoses, where specialists from various departments work together to make informed decisions.
Phase 3: Domain-Specific MoE Tuning
The final phase of Med-MoE’s development focuses on domain-specific Mixture of Experts (MoE) tuning. Domain-specific experts are selectively activated depending on the input, ensuring that only the most relevant experts contribute to the model’s decision-making. Additionally, a meta-expert is always activated to provide global context, further enhancing the model’s accuracy.
This selective activation of experts enables Med-MoE to operate efficiently with only a small number of active parameters, cutting computational costs by 30-50% compared to state-of-the-art models like LLaVA-Med.
Performance and Applications
The performance of Med-MoE has been rigorously tested across multiple medical datasets, including VQA-RAD, SLAKE, and PathVQA. These datasets consist of both open- and closed-ended medical visual question-answering tasks, as well as image classification challenges. Med-MoE has consistently outperformed or matched the best existing models while using far fewer parameters.
For example, the model demonstrated a notable improvement in zero-shot settings, achieving up to 9.4% better performance than LLaVA-Med in closed-ended VQA tasks.
Med-MoE also excelled in medical image classification tasks, such as PneumoniaMNIST and OrganCMNIST, proving its versatility in handling a wide range of medical data. Its success in these tasks underscores its potential as a practical tool in resource-limited healthcare settings, where computational resources are often scarce.
The model’s efficiency makes it highly deployable in real-world clinical environments, particularly those lacking access to high-end GPUs and computing power.
Conclusion
Med-MoE represents a significant advancement in the field of medical AI, combining cutting-edge machine learning techniques with practical utility. Its modular, expert-driven architecture allows it to handle both generative and discriminative tasks, all while reducing the computational burden traditionally associated with MLLMs.
By offering state-of-the-art performance with fewer parameters, Med-MoE paves the way for more accessible, scalable AI solutions in healthcare. Its potential to improve diagnostic accuracy and streamline clinical workflows makes it an essential tool for the future of medical AI.

[…] Read More! […]